Support Vector Machines (SVM)
Support vector machines are supervised learning models with associated learning algorithms that analyze data and recognize patterns, used for classification and regression analysis.
Gived an set of of training examples, each marked for belonging to one of two categories, an SVM training algorithm builds a model that assigns new examples into one category or the other, making it a non-probabilistic binary linear classifier.
An SVM model is a representation of the examples as points in space, mapped so that the examples of the separate categories are divided by a clear gap that is as wide as possible. New examples are then mapped into that same space and predicted to belong to a category based on which side of the gap they fall on.
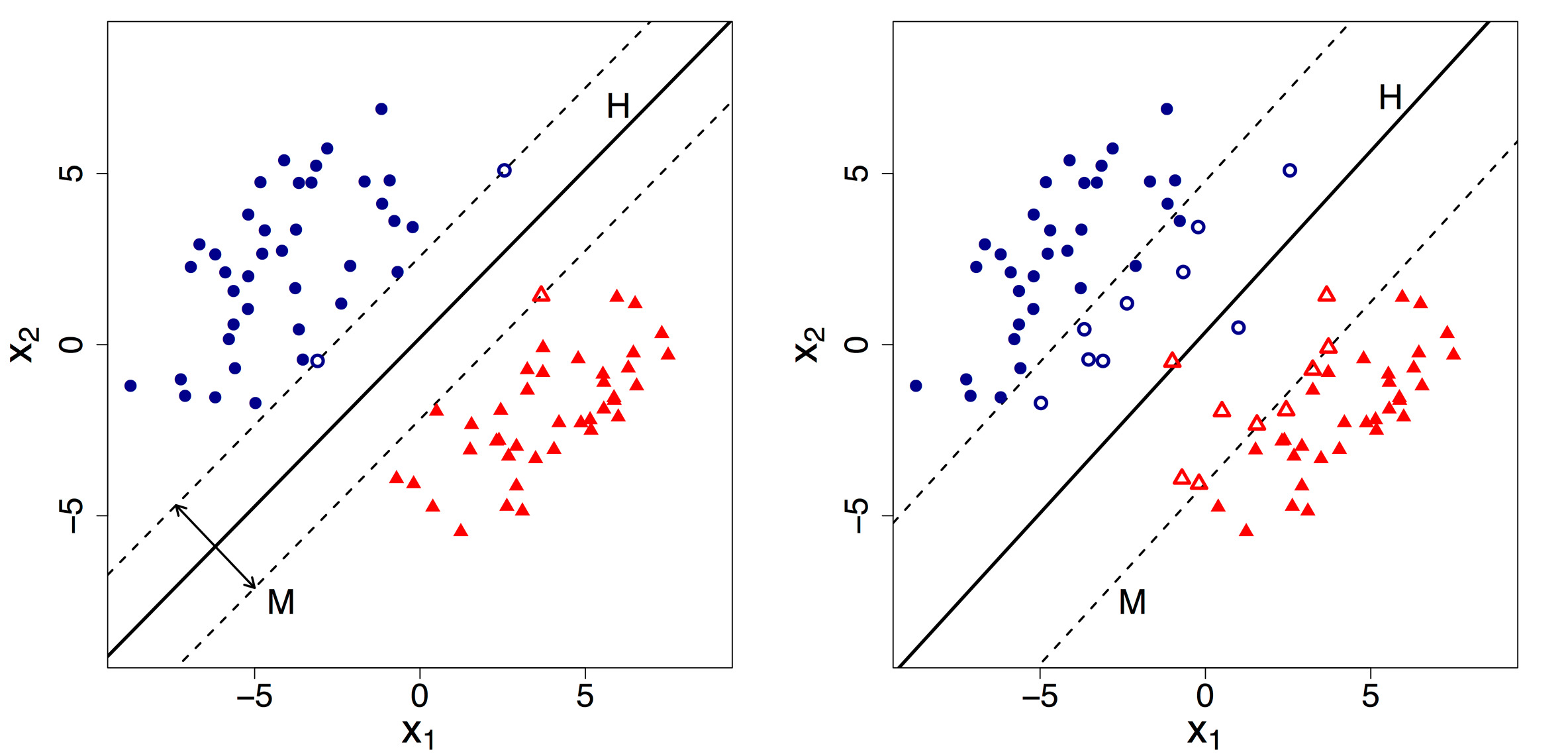
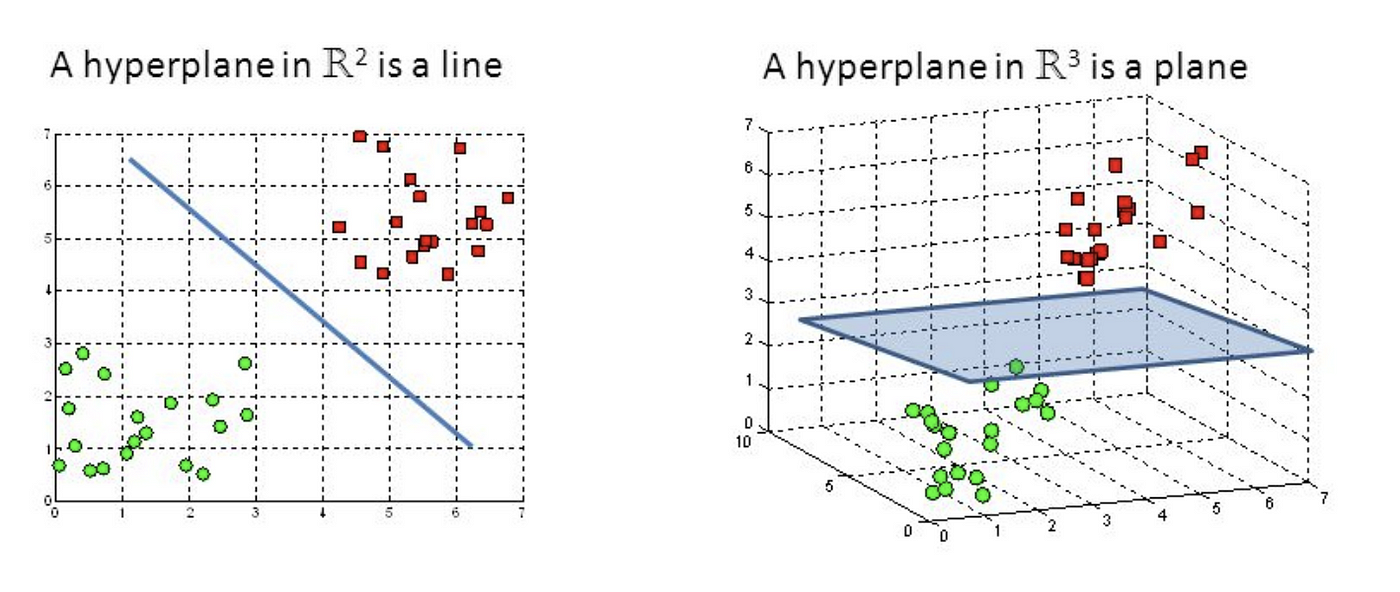
On a cartesian plan the SVM algorithm tries to find the best line that separates the two classes. This line is called hyperplane. The best hyperplane is the one that has the maximum margin, or distance between the two classes. The points that define the margin are called support vectors.
It's possisble to expand this idea to non-linearly separable data by using the kernel trick. The kernel trick is a method of using a linear classifier to solve a non-linear problem. It transforms the linearly inseparable data into linearly separable data by adding another dimension to it. The kernel trick is used to find a hyperplane in the high dimensional space that classifies the data points. The kernel function is what is used to transform the data into higher dimensions. The most common kernel functions are linear, polynomial, and radial basis function (RBF).
Python Implementation
Imports & Dataset
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
from sklearn.datasets import load_breast_cancer
cancert = load_breast_cancer()
print(cancer_dataset['DESCR'])
Preparing data
from sklearn.model_selection import train_test_split
X = pd.DataFrame(
cancer['data'],
columns=cancer['feature_names']
)
y = cancer['target']
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.3,
random_state=101
)
Support Vector Classifier
SVC is a type of SVM that supports multi-class classification. It does this by using the one-vs-one (OvO) method. OvO is a pairwise strategy, in which one classifier is trained for each pair of classes. If there are N classes in the problem, N * (N - 1) / 2 classifiers are constructed and each one trains data from two classes. At the end, the class that wins the most "votes" is the one that is predicted by the model.
It's a supervised learning algorithm that is commonly used for classification tasks. It belongs to the family of Support Vector Machines (SVMs). The SVC algorithm finds the optimal hyperplane in a high-dimensional feature space that maximally separates the different classes of data points. It achieves this by identifying a subset of training samples, called support vectors, which are crucial in defining the decision boundaries.
from sklearn.svm import SVC
svc_model = SVC()
svc_model.fit(X_train, y_train)
Predictions and Evaluations
predictions = svc_model.predict(X_test)
from sklearn.metrics import classification_report, confusion_matrix
print(confusion_matrix(y_test, predictions))
print(classification_report(y_test, predictions))
Gridsearch
Gridsearch is a method used to find the best combination of hyperparameters for a model. It does this by creating a grid of all the possible combinations of hyperparameters and then evaluating each one using cross-validation. The combination of hyperparameters that gives the best result is the one that is chosen.
Once the best combination of hyperparameters is found, the model is trained again using those hyperparameters and the final model is created.
from sklearn.model_selection import GridSearchCV
param_grid = {
# C controls the cost of misclassification on the training data.
# A large C value gives you low bias and high variance, because it penalizes the cost of misclassification a lot.
# A small C value gives you higher bias and lower variance.
'C': [0.1, 1, 10, 100, 1000],
# I'ts the gamma parameter of the RBF kernel, which is the kernel that is used in the SVM algorithm.
# The gamma parameter defines how far the influence of a single training example reaches.
# A high gamma value means that a single training example has a far reach, whereas a low gamma value means that the reach of a single training example is closer.
# In other words, a low gamma value considers a point to be similar to its neighbors, whereas a high gamma value considers a point to be similar to only its immediate neighbors.
# If gamma is too high, the algorithm will try to fit the training set very well, which will cause over-fitting.
# If gamma is too low, the model will not be able to capture the complexity (or “shape”) of the data set, which will cause under-fitting.
'gamma': [1, 0.1, 0.01, 0.001, 0.0001],
# Default kernel is 'rbf' (Radial Basis Function), which is a general-purpose kernel.
'kernel': ['rbf']
}
grid = GridSearchCV(
# use SVC (Support Vector Classifier) as estimator
SVC(),
# bind param_grid to constructor grid_search
param_grid,
# controls the verbosity: the higher, the more messages
verbose=3
)
Gridsearch Results
grid.fit(X_train, y_train)
# that's the best estimator found by GridSearchCV (SVC with C=1, gamma=0.0001 and kernel='rbf')
# this value had the best cross-validation score
grid.best_params_
# that's the best score found by GridSearchCV
# differently from best_params_, this value is the mean cross-validated score of the best_estimator
grid.best_estimator_
grid_predictions = grid.predict(X_test)
print(confusion_matrix(y_test, grid_predictions))
print(classification_report(y_test, grid_predictions))